To prosecute our goal of understanding the range of biological activities of complex mixtures with appropriate speed, breadth and precision, we are employing high innovative, orthogonal cell-based information rich phenotypic screening approaches developed and/or adapted in the MacMillan and Lokey laboratories.
Functional Signatures of Ontology
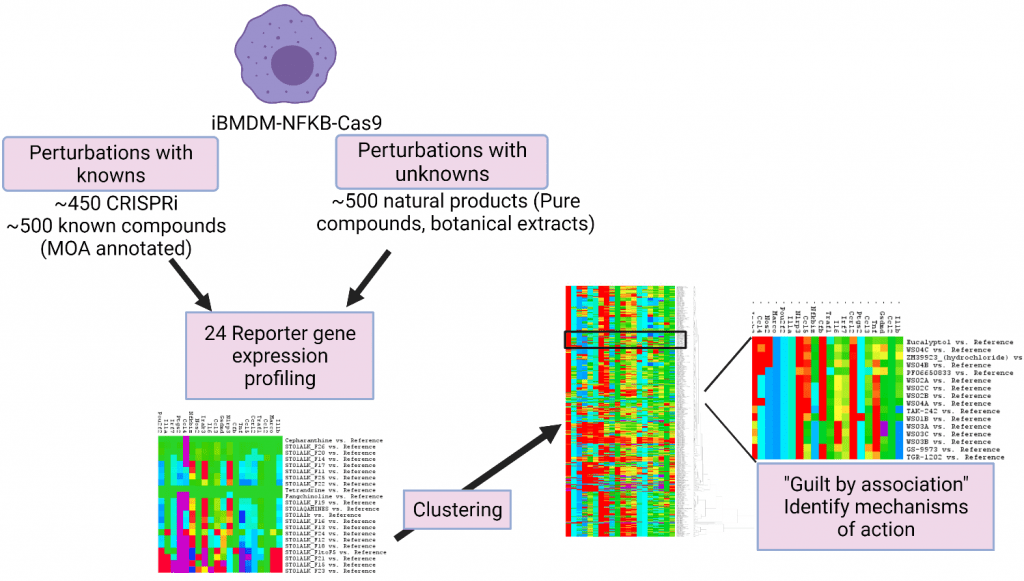
Functional Signatures of Ontology (FuSiOn) utilizes perturbation-induced gene expression signatures coupled with pattern-matching tools to produce verifiable guilt-by-association MOA hypotheses. The initial platform used the colon cancer cell line HCT116 and six reporter genes to generate expression signatures of “known” perturbagens such as siRNAs or miRNAs, for comparison to “unknown” perturbagens such as natural product fractions and pure natural products. This method has been used extensively to characterize the mechanism of pure natural products and generate profiles of complex botanical mixtures.
As part of this program we have made two important additions. 1) We have expanded the biological range of the FuSiOn approach by focusing on the use of primary bone marrow derived mouse macrophages in conjunction with the measurement of 14 specific genes. These measurements are made in the presence and absence of LPS, to look at a range of immunomodulatory activities. 2) we have adopted the robust and more prevalent Nanostring nCounter technology to make the FuSiOn approach more accessible to other groups.
Cytological Profiling
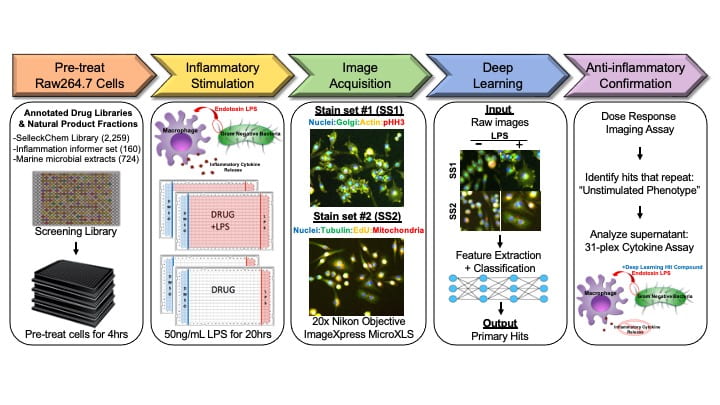
Cytological profiling (CP), utilizes high-content image analysis of perturbation-treated cells stained with a panel of fluorescent probes to extract sets of cytological features that are then used with pattern-matching tools to predict MOA. The initial CP platform utilized unsynchronized HeLa cells, which, after treatment with perturbagen are fixed and stained with probes for S-phase progression, total DNA, phospho-histone H3, tubulin and actin. A total of 251 unique cytological features are then extracted for each perturbagen from automated fluorescence microscopy images. Clustering compounds by their CP fingerprints has revealed both well-established associations among compounds with the same target or MOA, as well as novel or unexpected associations and unique phenotypes of natural products.
We have expanded our traditional CP approach to be used in macrophages, in the presence/absence of LPS, to look for immunomodulatory effects. The expansion to macrophages has required the development of new bioinformatic tools and the use of machine learning to understand the complex biological response.